sequence members within each group, we can optimize the alignment between
the groups, using two-dimensional DP. The resultant alignment, in turn, is
the starting point for the next alignment of a different pair of groups. Each
iteration that improves the alignment between two sequence groups will also
improve the global alignment.
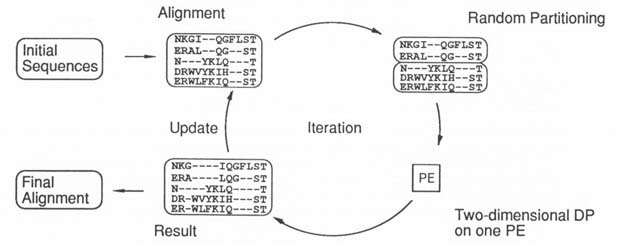
Figure 2 Original B-M algorithm |
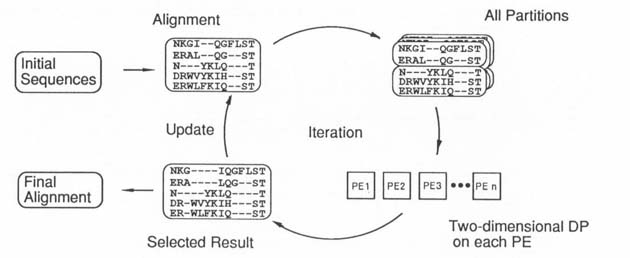
Figure 3 Parallel B-M algorithm |
This iterative strategy often results in much better multiple alignments
than those obtained by conventional algorithms. In Figure 4, (b) shows the
performance when this B-M algorithm is applied to seven sequences. Lines
(b-1), (b-2) and (b-3) are different in respect to random numbers. The quality
of the results is superior to that obtained by the tree-based algorithm, shown
as a horizontal line (a). However, the B-M algorithm needs a large amount of
time as the number of sequences grows.
We can reduce the execution time, when a parallel machine is available.
Figure 3 shows the algorithm of our parallel iterative aligner. Every possible
partitioning into two groups of aligned sequences can be respectively evaluated
by two-dimensional DP in a parallel way. In each iteration, the evaluation is
executed in parallel and the alignment which has the best score is selected as
the starting point for the next iteration. The performance of this method is
shown as line (c) in Figure 4. This shows that the parallel iterative aligner
performs better than the original B-M method in terms of execution time.
- 60 -


