1). A score is assigned to each arrow. We search a path from the top left
node to the bottom right node, maximizing the total score of the arrows. In
this case, the set of arrows that connect black circle nodes is the best path.
This best path corresponds to the optimal alignment.
Scores on arrows should reflect similarity between compared characters. In
the case of protein sequence alignment, Dayhoff's odds matrix is the most
popular way of obtaining the scores. The matrix was obtained by statistical
analysis of the mutation probability of amino acids.
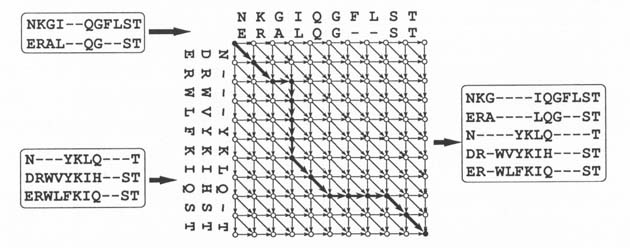
Figure 1 Two-dimensional DP |
Theoretically, N-dimensional DP provides an optimal alignment of N groups
of sequences. However, N-dimensional DP operates in exponential time as N
grows. When N is more than three, it does not complete in a realistic time
frame. Conventionally, researchers in the biological field make a multiple se-
quence alignment by merging groups of aligned sequences. A conventional
algorithm, called the tree-based algorithm, merges them in tree-like order us-
ing two-dimensional DP.
Though the execution time of the conventional algorithm is manageable, the
quality of its resultant alignment is not high enough yet. Thus, researchers
fairly often have to do multiple sequence alignments by hand. The large
number of sets of sequences to be aligned have become a burden on those
researchers.
Parallel iterative aligner
We developed a parallel iterative aligner, whose execution will be demon-
strated, in order to improve the quality of automatic multiple sequence align-
ment. The algorithm of this parallel iterative aligner is based on the Berger-
Munson algorithm. Firstly, we introduce the B-M algorithm, and then we
explain the parallel iterative aligner.
The B-M algorithm features a novel randomized iterative strategy so as
to generate a high-score multiple sequence alignment. Figure 2 illustrates
the iterative strategy, whose procedure is as follows: the initially aligned
sequences are randomly divided into two groups. By fixing the alignment of
- 59 -

