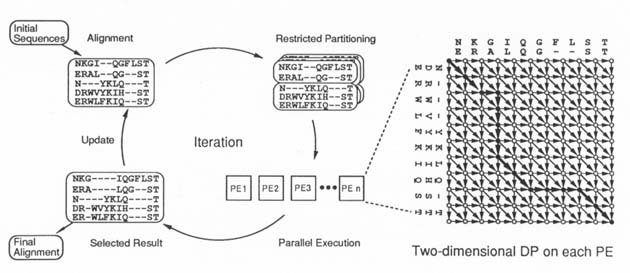
Operation of Parallel Iterative Aligner
ABSTRACT Every protein is a chain of twenty kinds of amino acids. Its structure and function are determined by the sequence of these amino acids. Protein sequence analysis is vital to the prediction of protein structure and function, and to the analysis of the evolutional relationship among proteins. We built the "Parallel Iterative Aligner" for this analysis using a PIM machine. It solves a typical protein sequence analysis problem, that of multiple sequence alignment. This kind of combinatorial problem generally requires a large amount of computation. Our Parallel Iterative Aligner, however, can quickly solve practical-sized problems, producing high-quality answers. KEY FEATURES Iterative Improvement The system partially improves temporary alignment in an iterative way, and can effectively reach high-quality alignment as a result. Speed-up by Parallel Execution Multiple branches of a search tree in this combinatorial problem are eval- uated in parallel by using many processing elements in each iteration. Pruning of Search Tree The heuristic method, "Restricted Partitioning Technique," prunes a large number of branches in the search tree and makes it possible to solve the combinatorial problem in a practical amount of time.
 Operation of Parallel Iterative Aligner |